Demonstration of a Superior Deep-UV Surface-Enhanced Resonance Ram an Scattering (SERRS) Substrate and Single-Base Mutation Detection in O ligonucleotides
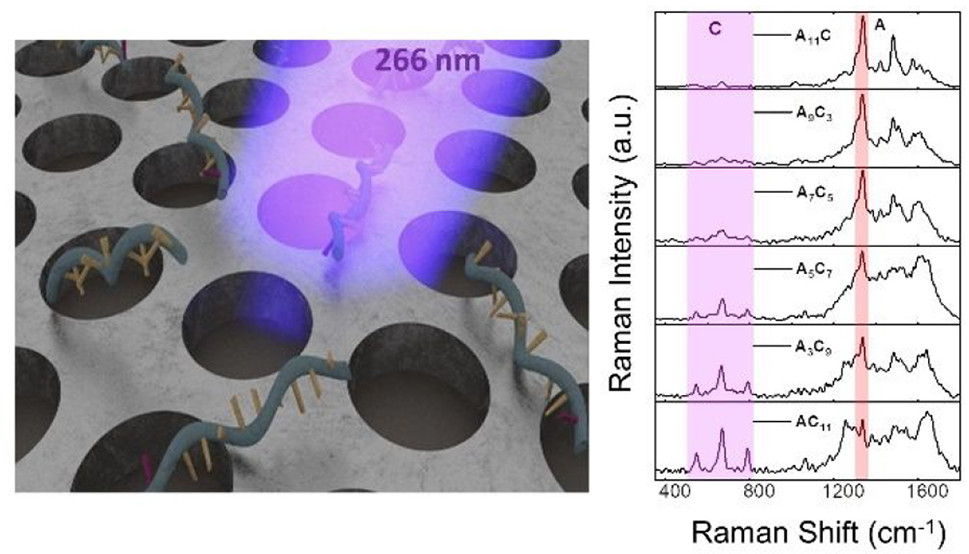

Raman spectroscopy identifies molecules based on their vibrational finger
print. However, its sensitivity is limited due to the small scattering cross-se
ction. To address this, SERS utilizes localized surface plasmons to enhance
the light-matter interaction by several orders of magnitude. By increasing
the excitation frequency to deep-UV(DUV) and introducing resonance eff
ects, the Raman signals can be significantly boosted.
Using finite difference time domain analysis, we determined the optimal d
iameter and periodicity of the Al plasmonic nanohole array to achieve LSP
R at 266 nm, matching the wavelengths of the incident laser and oligonucl
eotide absorption. The nanohole diameter was set at 100 nm for easier fabrication, while the periodicity was swept between 150 and 250 nm to opt
imize the LSPR at 266 nm. At the optimized periodicity of 200 nm, the Al p
lasmonic nanohole array exhibited a reflectance minimum at 266 nm. Imp
ortantly, this optimized design not only simplified fabrication but also res
ulted in a high density of hot spots in the laser excitation area, further enh
ancing the sensitivity and performance of the substrate.
We epitaxially grow an Al film on a sapphire(Al2O3) substrate, by using a
plasma-assisted molecular beam epitaxy(PA-MBE) method. We introduce
an optimized plasmonic nanohole array by using electron beam lithograp
hy and a reactive ion etching process. We measure the reflectance spectra
by using a micro-DUV reflectance setup to confirm the behavior of the pla
smonic resonance. Both the experiment and simulations overlap with the
reflectance dip at around 266 nm, which confirm the designated plasmoni
c resonance of the fabricated Al nanohole array.
To evaluate the substrate's performance, we detected five nucleotides(A,
T, C, G, U) using the Al plasmonic nanohole array. The substrate exhibited t
he highest enhancement factors (EF) of up to ~10^6.This showcases the p
otential of DUV-SERRS substrates for sensitive molecular analysis.
National Tsing Hua University (NTHU), established in 1911 and located in Hsinchu, Taiwan, is one of the top research universities in the country. NTHU offers a wide range of programs in fields such as engineering, science, management, and humanities. The university is known for its strong emphasis on innovation, research excellence, and fostering global perspectives. With a commitment to academic rigor and interdisciplinary collaboration, NTHU plays a key role in advancing knowledge and technological development, contributing to both Taiwan’s growth and the global academic community.
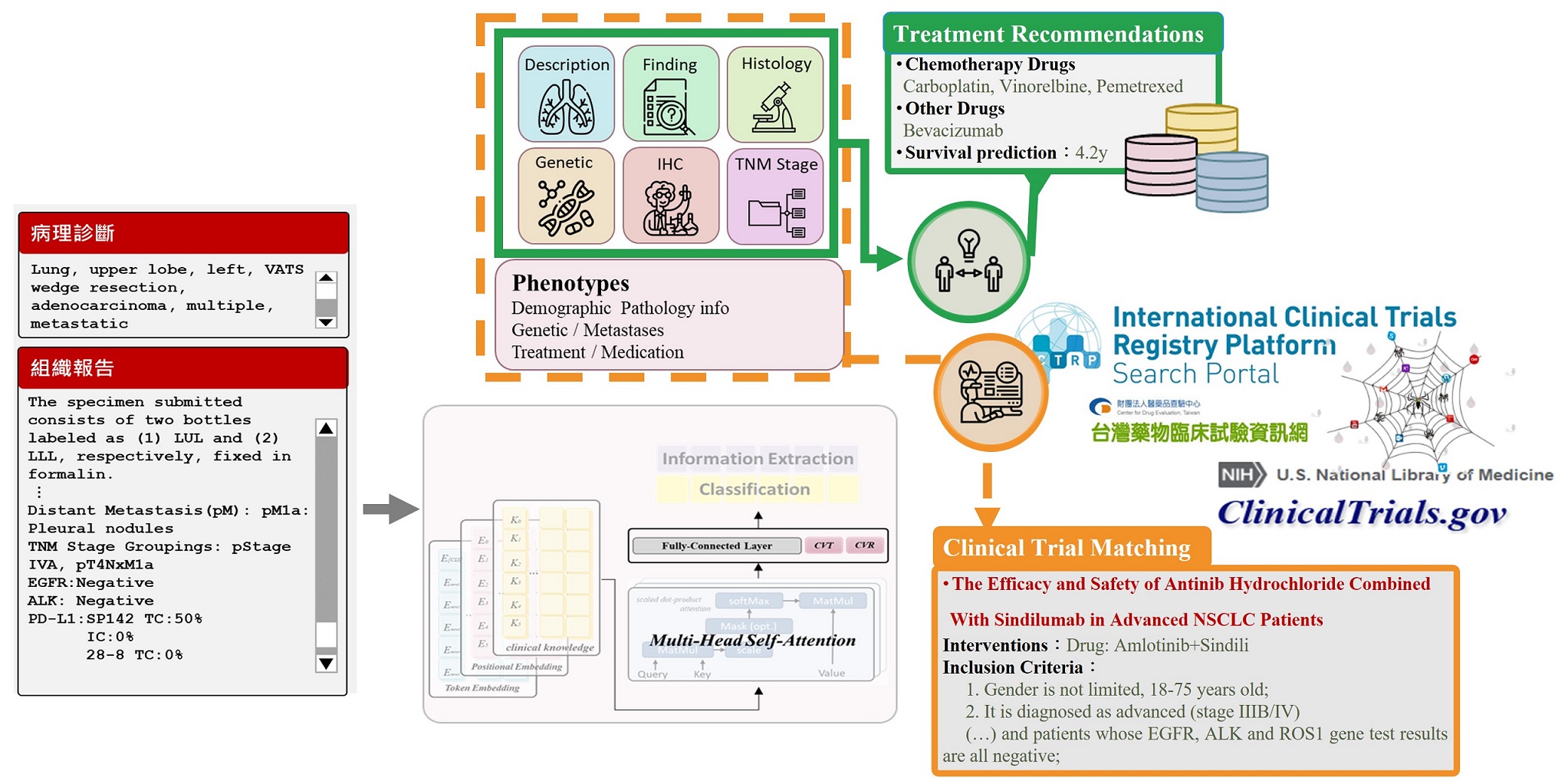
Using Generative Deep Learning to Predict Drug Response and Survival, a nd Automatically Match Clinical Trials for Advanced Lung Cancer.
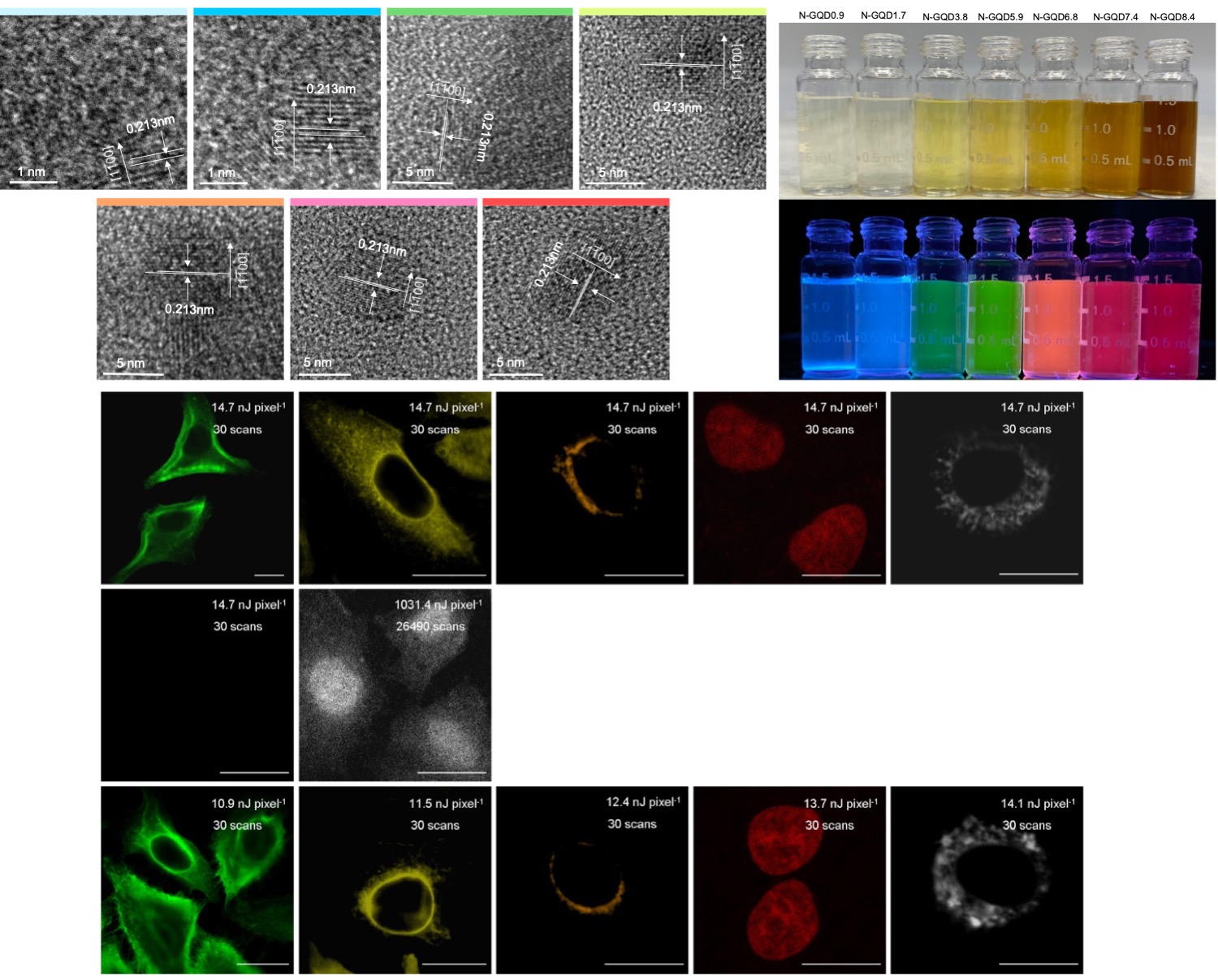
The Application of Multi-Functional Graphene-Based Materials in Targeti ng Detection, Diagnosis and Therapy
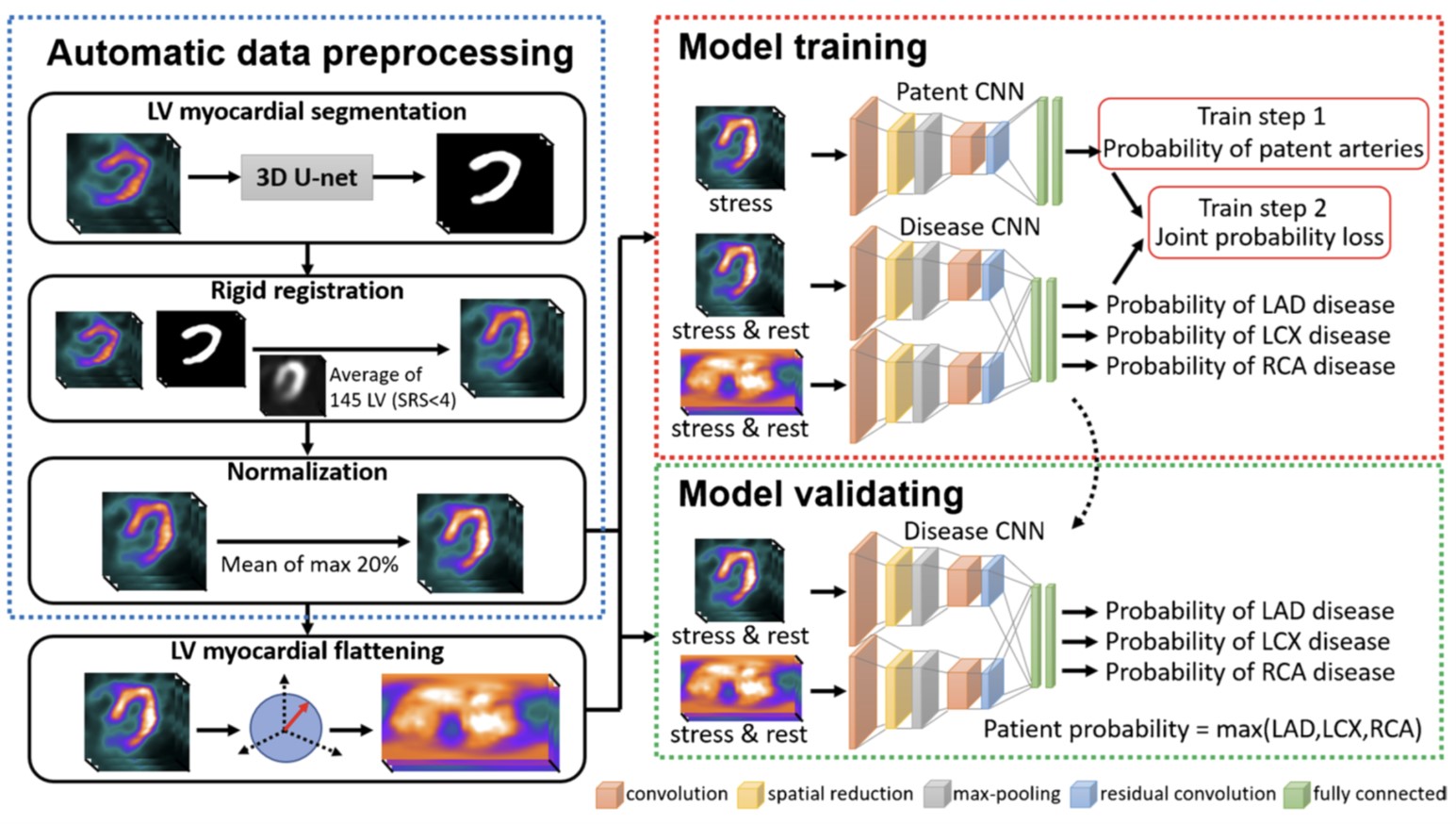
One Model Fit All: Revolutionary One-Stop Shop System for Predicting Co ronary Stenosis without Normal Database in Myocardial Perfusion Imagin g
Quantitative Analysis of Drug Toxicity Using Metallic Nanostructures and Contrast-Enhanced Surface Plasmon Imaging Technology
Technology maturity:Experiment stage
Exhibiting purpose:Display of scientific results
Trading preferences:Negotiate by self
*Organization
*Name
*Phone
*Main Purpose
*Discuss Further
*Job Category
*Overall Rating
*Favorite Area
*Key Tech Focus
*Willing to Receive Updates?
Other Suggestions
Coming soon!